Projects
RNA and Protein Atlas for Human
Bacterial Pathogens
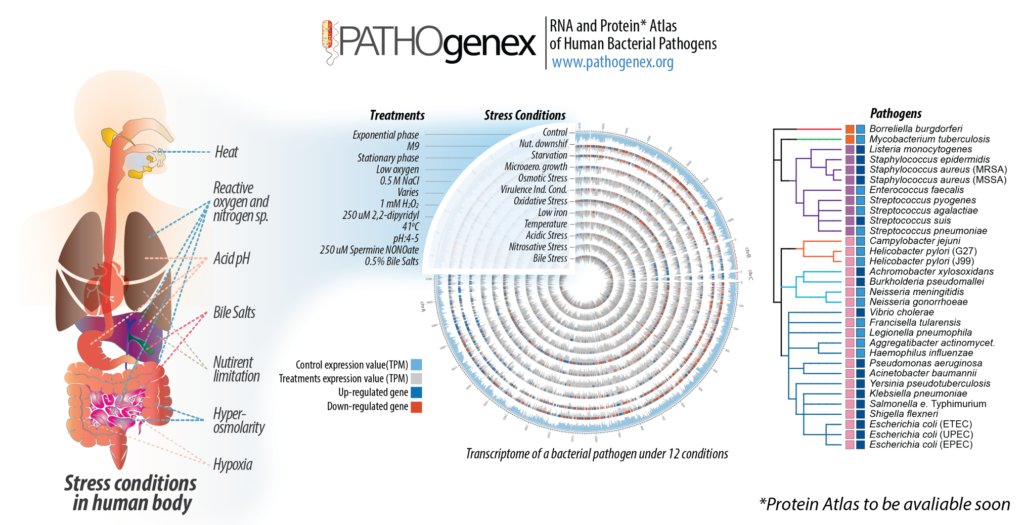
Bacterial processes necessary for adaption to stressful host environments are potential targets for new antimicrobials. The potential relevance of the in vitro stress conditions and responses is supported by comparisons with available in vivo transcriptomes of clinically important pathogens. Here we analyzed global expression profiles of 32 different bacterial pathogens under 11 infection-relevant stress conditions. Data were collected in an interactive RNA atlas, named PATHOgenex.
Single Cell Transcriptomics
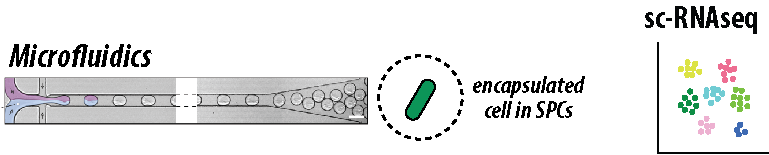
Bacterial populations exhibit significant heterogeneity, allowing distinct cell subgroups to thrive in diverse environments. This environmentally organized heterogeneity prompts bacteria to respond to dynamic changes through unique transcriptional programs, posing substantial challenges in assaying individual cells uniformly. Recent advancements in single-cell techniques, particularly single-cell RNA sequencing (scRNA-seq), offer a powerful means to unravel cellular heterogeneity during infections and discern subpopulation variations that influence individual cell outcomes. However, current bacterial scRNA-seq methods suffers from low number of cells being sequenced and low number of transcripts detected in each cell due to leakage of nucleic acids due to mechanical disruption of cell wall during permeabilization for subsequent enzymatic reactions. Therefore, we are developing a novel bacterial scRNA-seq method by using advantage of semi-permeable hydrogel microcapsules (SPCs), which allows encapsulation of bacterial cells by microfluidics device and multiple enzymatic reactions once the cells are lysed inside SPCs.
Prosthetic Joint Infection
Human bacterial pathogens are capable of adapting to a range of stressful conditions that they encounter within their host environment. This adaptive ability enables them to thrive in hostile environments and even develop complex forms of colonization such as biofilm formation, which can be challenging to treat. This adaptive ability enables them to survive in hostile environments and even develop complex forms of colonization such as biofilm formation, which can be challenging to treat. We planned to develop novel methods to detect culturable and non-culturable bacterial species to reach complete bacterial transcriptomes from PJIs. All the complex and rich datasets will be processed to uncover unknown and vital information by employing systems biology, which includes mathematical and computational disciplines, and artificial intelligence algorithms. The primary objective is to identify bacterial determinants that can be targeted for developing new antimicrobials by unraveling critical mechanisms and/or determinants that enable bacteria to maintain infections in humans.